Extraktion von Plasmid-DNA aus bakteriellen Lysaten
Beschreibung: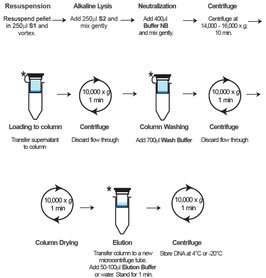
Das GF-1 Plasmid DNA Extraction Kit ist für eine schnelle und effiziente Reinigung von Plasmid-DNA aus bakteriellen Lysaten konzipiert.
Dieses Kit nutzt die alkalische Lyse-SDS-Methode, um Zellen zu lysieren und Plasmid-DNA freizusetzen.
Spezielle im Kit enthaltene Puffer optimieren die Bindung der DNA auf eine spezielle Glasfilter-Membran zur effizienten Gewinnung von hochreiner Plasmid-DNA. Die hochreine Plasmid-DNA kann für alle Arten von Folgeexperimenten in der Molekularbiologie, wie zum Beispiel Restriktionsenzymverdau, radioaktive/fluoreszierende DNA-Sequenzierung, PCR, Ligation und Transformation weiterverwendet werden.
Features:
– Gewinnung von bis zu 20µg DNA
– Mehrere Proben können in weniger als 30 Minuten verarbeitet werden
– Keine organisch-basierte Extraktion nötig
Anwendung: Für die Isolierung von Plasmid-DNA
Kit Bestandteile:
– GF-1 columns
– Collection tubes
– Lösung 1 (S1)*
– Lösung 2 (S2)*
– Neutralisationspuffer (Puffer NB)
– Waschpuffer (Konzentrat)*
– RNase A (DNase-frei)*
– Elutionspuffer
– Handbuch
* Bitte beachten Sie Verdünnung von Lösungen und Lagerung und Stabilität vor der Verwendung dieses Kits.
| Artikel | Beschreibung |
| PL05 | Plasmid DNA Extraction Kit The Plasmid DNA Extraction Kit is designed for rapid and efficient purification of high copy and low copy plasmid DNA from bacterial lysates. St./Karton: 5 preps |
| PL50 | Plasmid DNA Extraction Kit The Plasmid DNA Extraction Kit is designed for rapid and efficient purification of high copy and low copy plasmid DNA from bacterial lysates. St./Karton: 50 preps |
| PL100 | Plasmid DNA Extraction Kit The Plasmid DNA Extraction Kit is designed for rapid and efficient purification of high copy and low copy plasmid DNA from bacterial lysates. St./Karton: 100 preps |
| PL200 | Plasmid DNA Extraction Kit The Plasmid DNA Extraction Kit is designed for rapid and efficient purification of high copy and low copy plasmid DNA from bacterial lysates. St./Karton: 200 preps |

MSDS
Referenzen
Tohidi, F., et al. (2016) Development of a Novel In Vitro Assay for the Evaluation of Integron DNA Integrase Activity.. Biotechnology & Biotechnological Equipment11(57), Taylor and Francis Online, p.1-7.
Uttatree,S., Charoenpanich, J. (2016) Isolation and characterization of a broad pH- and temperature-active, solvent and surfactant stable protease from a new strain of Bacillus subtilis. . Biocatalysis and Agricultural Biotechnology. 8. Pp.32-38
Hamidinejat, H., et al. (2015) Development of an Indirect ELIS using Different Fragments of Recombinant Ncgra 7 for Detection of Neospora caninum Infection in Cattle and Water Buffalo. BIran Journal of Parasitology. 10(1), p.69-77.
Hesampour, A., et al. (2014) Comparison of Biochemical Properties of Recombinant Phytase Expression in the Favorable Methylotrophic Platforms of Pichia pastoris and Hansenula polymorpha. Progress in Biological Sciences. 4(1), p. 97 – 111.
Gul, R., et al. (2012) Expression and Sequence Characterization of Growth Hormone Binding Protein of Nili-Ravi Buffaloes (Bubalus bubalis). African Journal of Biotechnology. ProQuest. 11(57), p. 12103-12109
Gul, R., et al. (2012) Expression and Sequence Characterization of Growth Hormone Binding Protein of Nili-Ravi Buffaloes (Bubalus bubalis). African Journal of Biotechnology. ProQuest. 11(57), p. 12103-12109
Pongsilp, N., et al (2012) Genotypic Diversity among Rhizospheric Bacteria of Three Legumes Assessed by Cultivation-dependent and Cultivation-independent Techniques. World Journal of Microbiology and Biotechnology.ProQuest. 28, p. 615-626
Charoenpanich, J., Suktanarag, S., & Toobbucha, N. (2011)Production of Thermostable Lipase by Aeromonas sp. EBB-1 Isolated from Marine Sludge in Angsila, Thailand ScienceAsia37: 105-114.
Uttatree, S., Winayanuwattikun, P., & Charoenpanich, J. (2010) Isolation and Characterization of a Novel Thermophilic-organic Solvent Stable Lipase from Acinetobacter baylyi Applied Biochemistry and Biotechnology.
